
The aim of this review is to provide an updated approach to this challenge. The estimate of variant impact on RNA processing is not always simple, and can lead to improper variant classification. In addition, the presence of genomic variants, involving consensus splicing regulatory sequences in different parts of a gene, may modify the splicing process, alter the mRNA and eventually affect the corresponding protein-coding sequence. Regulation of alternative splicing is complex with several elements interacting in a coordinated manner including cis-acting and trans-acting factors, spliceosome components as well as chromatin or RNA structure together with the presence of alternative transcription initiation (ATI) or alternative transcription termination (ATT) sites. During AS, exons, or portions of exons or noncoding regions within a pre-messenger RNA (pre-mRNA) transcript, are differentially fixed or skipped, resulting in multiple protein isoforms. More than 95% of human genes have been found to undergo alternative splicing in a developmental, tissue-specific or signal transduction-dependent way. AS is crucial to enhance gene expression, to drive cellular differentiation and organism development. This phenomenon can be possible thanks to mechanisms of alternative splicing (AS), a process that was first proposed by Gilbert in 1978. This exorbitant number is nothing when compared with the almost 90,000 different proteins that form human proteome. How many protein coding genes have been described in humans? The answer is approximately 25,000–30,000. For both prediction and validation steps, benefits and weaknesses of each tool/procedure are accurately reported, as well as suggestions on which approaches are more suitable in diagnostic rather than in clinical research. Next, we report the experimental methods to validate the predictive analyses are defined, distinguishing between methods testing RNA (transcriptomics analysis) or proteins (proteomics experiments). First, we review the main computational tools, including the recent Machine Learning-based algorithms, for the prediction of splice site variants, in order to characterize how a genomic variant interferes with splicing process. Then, we provide an updated approach to improve splice variants detection. In this review, we introduce the key steps of splicing mechanism and describe all different types of genomic variants affecting this process (splicing variants in acceptor/donor sites or branch point or polypyrimidine tract, exonic, and deep intronic changes).
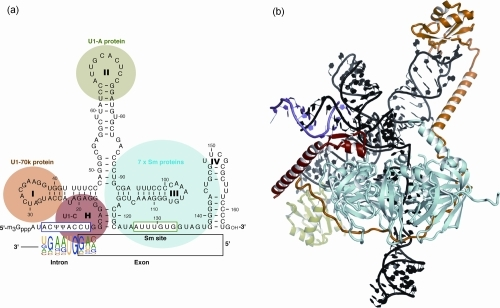
Any alteration (e.g., nucleotide substitutions, insertions, and deletions) involving consensus splicing regulatory sequences in a specific gene may result in the production of aberrant and not properly working proteins. Interestingly, more than 95% of human genes undergo AS, producing multiple protein isoforms from the same transcript.

Alternative splicing (AS) is a crucial process to enhance gene expression driving organism development.


 0 kommentar(er)
0 kommentar(er)
